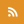High-resolution, 3D images of human chromosomes in single cells reveal how chromatin structure might influence its function
By Caitlin McDermott-Murphy
Remember those wonky Xs that textbooks teach are chromosomes? Those cartoons were science’s best guess at what chromosomes look like.
Now, Xiaowei Zhuang, the David B. Arnold, Jr. Professor of Science, and four Ph.D. candidates in the Department of Chemistry and Chemical Biology, Department of Physics, Department of Molecular and Molecular Biology and Biophysics Program in the Graduate School of Arts and Sciences— Bogdan Bintu, Seon Kinrot, Jun-Han Su, and Pu Zheng—captured high-resolution 3D images of human chromosomes, the intricately organized houses for a human’s entire instruction manual—their DNA.
Extended in a straight line, the DNA inside a single cell can reach the length of an average bicycle—about six feet. But the 23 pairs of chromosomes inside each cell nucleus are like obsessive-compulsive packrats: They wind genetic material in tight, complex structures. For a human—or any organism—to grow and function properly, cells must constantly divide to replace old, worn-out cells with new ones. A hitch in the chromosome structure could lead to an error in gene expression, which could cause disease.

Chromosomes, often pictured as an "X" as seen here on the right, are made up of DNA wrapped in complex formations around histone proteins. Now, the Zhuang lab captured images of how this structure forms and how errors impact function. Image Credit: National Institutes of Health
That’s why Zhuang and her team wanted a closer look at how chromatin—the DNA and associated proteins that make up each chromosome’s complex architecture—is structured and how that structure influences function. In a paper published in Cell, they report a new way to image chromatin structure and function at the same time, capturing both a 3D bird’s eye view of all 46 chromosomes and a close-up of just two, to start piecing together how and why one influences the other.
To create their 3D chromosomal map, the team first needed to pinpoint the location of genomic loci—dots along the chromatin structure. By connecting the dots—a lot of dots—they could form a fairly comprehensive picture.
But there was a snag. Previously, the number of loci one could simultaneously image and identify was traditionally limited to the number of color channels that could be discerned in a fluorescence microscope, which is no more than a handful. If there are only a few different hues, only a few loci can be captured in one image, which can’t make a comprehensive picture.
So, Zhuang and her team came up with a multiplexed approach: Image a few different loci, quench the signal, and then image a few more in rapid succession. With that technique, each locus gets two identifying marks: color and image round. With that approach, they could image, localize and identify several tens to hundreds of loci in one go.
Still, for a 3D image of the genome, they needed more—thousands—so they turned to a language often used to organize huge amounts of information: binary codes. By imprinting binary barcodes on different genomic loci, they could image far more loci at a time. For example, a molecule imaged in round one but not round two gets a barcode starting with “10.” With ten-bit barcodes, the team could differentiate more than 1,000 loci in just ten rounds of single-color imaging or five rounds of two-color imaging. They also built error-detection into their barcodes, which increased the number of imaging rounds needed but achieved high detection accuracy. Even so, with just a few tens of rounds, they imaged and localized the thousands of loci needed for a comprehensive picture.

The Zhuang lab used both color and image round to image and localize thousands of loci. Image courtesty of the Zhuang lab
“In this combinatorial way, we can increase the number of loci imaged and identified much more rapidly,” said Zhuang. She and her lab developed this approach, called MERFISH (multiplexed error robust fluorescence in situ hybridization), to image the transcriptome, the full suite of messenger RNA.
Now, the team trained MERFISH to capture the 3D genome, imaging about 2,000 chromatin loci per cell, or enough to form a high-resolution image of what the structure looks like in its native habitat. But they didn’t stop there: They also imaged transcription activity (when RNA replicates genetic material from DNA) of more than 1000 genes and nuclear structures, including nuclear speckles and nucleoli. With their 3D Google Maps of the genome, they can start to analyze how structure regulates genome function.
Researchers already know chromatin has different compartments and, within those, different domains, but what these terrains look like from cell to cell and how they function has been largely unknown. With their high-resolution images, the team determined that compartments of active chromatin (gene-rich chromatin prone to transcription) tend to flock to other active chromatin compartments. The same is true for inactive chromatin compartments, though active chromatin compartments hunt for each other over greater distances. They also found that the local chromatin environment impacts transcription activity: Genomic loci residing in an environment enriched with active chromatin tend to have higher transcription activity. Structure does influence function.
The team also discovered high variability between domains even in cells that are, otherwise, functionally similar. Even in their cell cultures—which were full of identical cells—no two chromosomes looked the same. Since these differences increase in the huge variety of cells that make up the human body, far more work needs to be done to build a Google Maps for all cell types.
“It's not going to be possible to build just on our work,” Zhuang said. “We need to build on many, many labs’ work in order to have a comprehensive understanding.” That’s why Zhuang not only shares instructions for other labs to replicate their experiments, she also offers trainings. There are too many biological mysteries for one group to solve.
What about the mystery of those oblong Xs in biology textbooks? Do chromosomes really look like that? Only in a small fraction of the time. Most of the time, they twist into other shapes to suit their function. Now, the Zhuang lab has the pictures to prove it.