David Liu presents two new CRISPR tools: CAMERA and xCas9
By Caitlin McDermott-Murphy
David Liu earned the title “Gene Corrector” for good reason. A professor of chemistry and chemical biology at Harvard University and core institute member of the Broad, Liu is also a vanguard pioneer of CRISPR-Cas9 gene-editing technology.
In recent years, Liu has earned recognition for his major advances to improve CRISPR’s versatility and accuracy. In 2016, he and postdoc Alexis Komer reported CRISPR’s first base editor, a technique that makes single-letter changes in the genome (e.g., C•G to T•A or A•T to G•C). Researchers already employ the tool in wheat, zebrafish and mice to treat genetic diseases, grow climate-resilient crops, and develop designer materials, foods, and drugs.
Now, in back-to-back papers, Liu presents two new CRISPR upgrades. The first, published in Science online, debuts CAMERA, a cellular “black box.” The second, published in Nature, unveils xCas9, a gene-editor that improves aspects of the widely used SpCas9.
Cellular 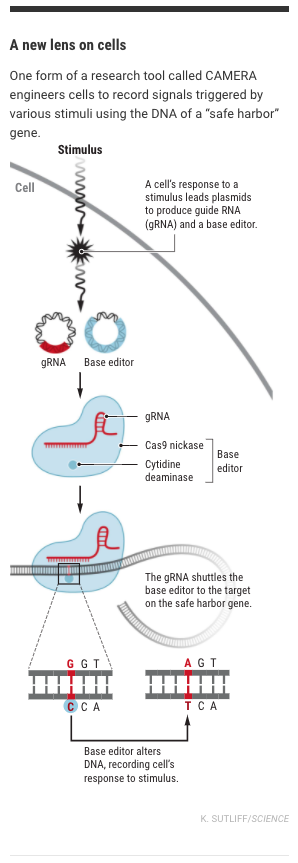 Detectives: CAMERA 1 and 2
Detectives: CAMERA 1 and 2
A good doctor, presented with an inconclusive case, will chase diagnostic clues in a patient’s environment, family medical history, and more. Cell biologists, likewise, have pursued methods to record moments in a cell’s life in order to understand its future. Armed with a log of intimate cellular data, researchers could unravel how cells transition from stem to muscle, neuron, and other cell types or from normal, healthy cells to cancer. This, in turn, could lead to greater understanding of disease progression and aging.
Now, Liu and postdoc Weixin Tang have enlisted CRISPR to assist. Their “CRISPR-mediated analog multievent recording apparatus (CAMERA) systems” harness the prolific Cas9 protein to record cellular data on DNA. The tools capture “multiple stimuli in bacteria or mammalian cells, including exposure to antibiotics, nutrients, viruses, light, and changes in Wnt signaling.” This last—Wnt signaling—figures in both embryo and cancer development.
CAMERA works in samples that contain as few as 10 cells and records event duration, simultaneous triggers, and can even “reset” to baseline to record anew. The work “is really beautiful stuff,” according to Timothy Lu, who developed a similar system at the Massachusetts Institute of Technology. He concedes that CAMERA offers “a level of efficiency and precision that goes beyond what we did earlier.”
Both systems use base editors and Cas9 nucleases to record, but they employ two distinct techniques.
CAMERA 1 co-opts the “plasmids” of DNA present in bacterial cells to document activity. The team designed two “recorder” plasmids, R1 and R2, whose population remain analogous due to the bacteria’s fierce regulation. Then, they insert a third plasmid, armed with CRISPR’s tools. This third provokes the cell to attack R1 when the cell experiences a certain stimulus, e.g., antibiotics or light. After exposure, the ratio of R1 to R2 plasmids signifies the duration of the experience.
The surreptitious CAMERA 2 acts more like a cellular spy. A modified Cas9 delivers its “guide” RNA (gRNA) into a gene whose DNA can be altered without harming the cell. From this vantage point, the tool records the presence and duration of specific stimuli onto their borrowed gene.
Though promising, Liu cautions against inflation of the tool’s capabilities. “The CAMERA systems are likely to be first used in research settings to illuminate cellular processes and signaling events,” he explained. “In principle, one might eventually use CAMERA-like systems to record changes in a patient’s cells, but such an application would require quite a lot (years) of additional development work.”
So, in the future, a good doctor might be able to review our cellular histories to provide accurate predictions about our future health. But such individualized treatment may yet be years or decades away.
CAMERA records both internal and external chatter that influence a cell
SOURCE: Russell Kightley/Science
A new, improved Cas9
Despite a wealth of recent advances and publicity, CRISPR remains imperfect.
The most popular CRISPR enzyme (spCas9 from the bacterium Streptococcus pyogenes) relies on specific DNA markers to pinpoint where to cut or edit. spCas9 scans the genome for stretches of DNA that end with three specific bases: N, where N denotes any of DNA’s four bases, followed by two guanines (Gs). Only about 1 in 16 DNA sites match this requirement.
Because of its restricted range, SpCas9 cannot reach a number of genes associated with human disease. And, it can confuse similar DNA stretches, resulting in risky “off-target” edits.
Now, Liu has engineered a modified version of the CRISPR-Cas9 enzyme. Using his rapid phage-assisted continuous evolution (PACE) system, his lab designed altered versions of spCas9 and narrowed in on those that could find a wider range of DNA stretches (known as protospacer adjacent motifs, or PAMs).
Named extended-PAM Cas9s or xCas9s, these laboratory-produced enzymes are both more versatile and precise. They accept more DNA sites—at least 1 in 4 instead of 1 in 16—and result in far fewer “off-target” edits. So, even though xCas9 can target more genes, it’s more discriminating than its predecessor.
"The study thus demonstrates that laboratory evolution can be used to address one of the biggest limitations facing the use of nature’s SpCas9—the PAM requirement—without sacrificing, and indeed while improving, DNA specificity"- Professor David Liu |
“The study thus demonstrates,” Liu notes, “that laboratory evolution can be used to address one of the biggest limitations facing the use of nature’s SpCas9—the PAM requirement—without sacrificing, and indeed while improving, DNA specificity.”
When asked for an explanation for this welcome, though unexpected, improvement, Liu simply says “the short answer is, ‘I don’t know.’ But it’s tempting to speculate that nature’s Cas9 evolved to be promiscuous enough to defend against mutating viral DNA, the natural target of Cas9, and that enough laboratory evolution can erode this promiscuity and improve DNA specificity.”
Although he and his lab will continue to test the efficacy of xCas9, Liu hopes that the new enzyme “with its broader DNA targeting scope and higher DNA specificity will become a preferred form of Cas9 for a wide range of genome editing and epigenome editing applications.” If so, CRISPR’s reach will continue to expand, promising future intervention for a wider range of genetic diseases.